How to configure plugins
There are two ways to configure the plugins:
1. Download PhyloSuite with plugins
When using Windows and Mac OSX systems, you can download PhyloSuite_xxx_Win32/64_with_plugins.rar and PhyloSuite_xxx_Mac_with_plugins.zip from https://github.com/dongzhang0725/PhyloSuite/releases, respectively. All plugins, except Python 2.7, Perl 5, Java, Rscript, HmmCleaner and trimAl (MAC) are included.
2. Configure plugins separately
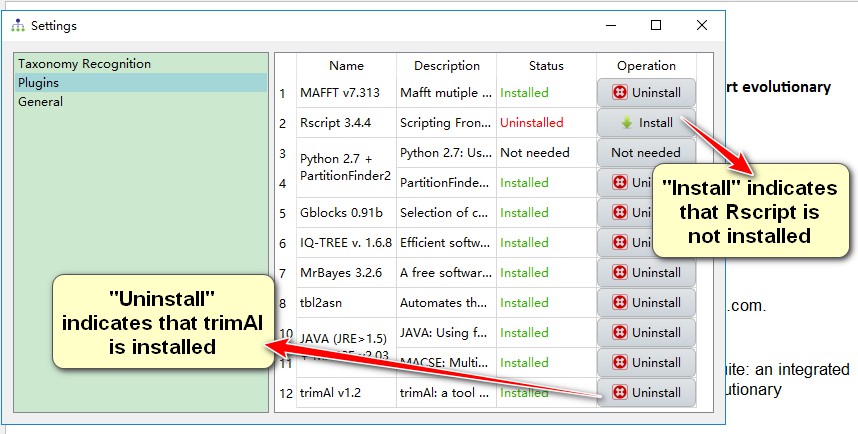
If Python 2.7, Perl 5, Java (JRE > 1.5), HmmCleaner.pl and trimAl have been installed and added to the environment variable ($PATH), they will be automatically detected by PhyloSuite. Open the plugin settings window via Settings-->Settings in the menu bar. There are two ways to configure your plugins:
2.1. Specify executable file
When you have the plugins already installed on your computer, you can just specify the paths directly.
- Click the
Installbutton of each plugin to open the configuration window; - In the
Specifybox, clickAddbutton to select files; - Select the executable file (see the table below);
- Select
Open; - Click
Ok.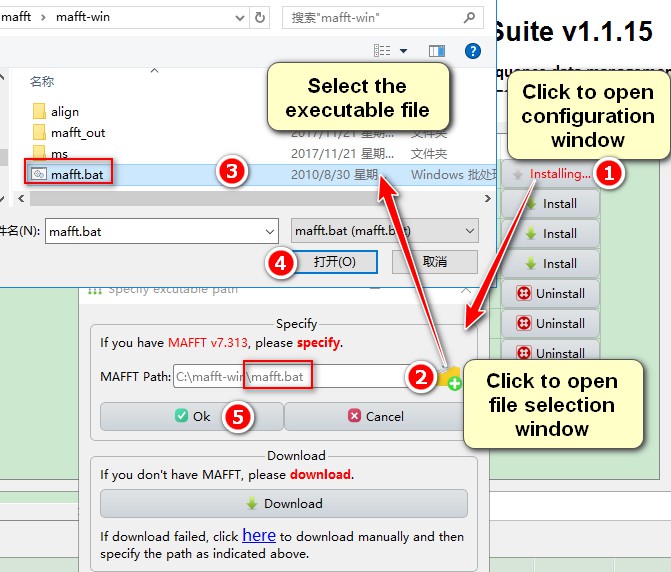
Programs | Executable File (Win) | Executable File (MAC) | Executable File (Linux)
——– | — | — | — | —
MAFFT v7.313 | path-to-MAFFT/mafft-win/mafft.bat | path-to-MAFFT/mafft-mac/mafft.bat | path-to-MAFFT/bin/mafft or
/usr/bin/mafft
Java (JRE > 1.5) | path-to-Java/java.exe | path-to-Java/java or usr/bin/java | path-to-Java/java or usr/bin/java
MACSE v2.01b | path-to-MACSE/macse_v2.03.jar | path-to-MACSE/macse_v2.03.jar | path-to-MACSE/macse_v2.03.jar
IQ-TREE v. 1.6.8 | path-to-IQ-TREE/bin/iqtree.exe | path-to-IQ-TREE/bin/iqtree | path-to-IQ-TREE/bin/iqtree
MrBayes 3.2.6 | path-to-MrBayes/mrbayes_x64.exe or
path-to-MrBayes/mrbayes_x86.exe | path-to-MrBayes/mb | path-to-MrBayes/bin/mb
PartitionFinder2 | path-to-PartitionFinder2/partitionfinder-2.1.1 | path-to-PartitionFinder2/partitionfinder-2.1.1 | path-to-PartitionFinder2/partitionfinder-2.1.1
Gblocks 0.91b | path-to-Gblocks/Gblocks.exe | path-to-Gblocks/Gblocks | path-to-Gblocks/Gblocks
trimAl | path-to-trimAl/trimal.exe | path-to-trimAl/trimal or usr/bin/trimal | path-to-trimAl/trimal or usr/bin/trimal
Perl 5 | path-to-Perl/perl.exe | path-to-Perl/perl or usr/bin/perl | path-to-Perl/perl or usr/bin/perl
HmmCleaner.pl | path-to-HmmCleaner/HmmCleaner.pl | path-to-HmmCleaner/HmmCleaner.pl or usr/bin/HmmCleaner.pl | path-to-HmmCleaner/HmmCleaner.pl or usr/bin/HmmCleaner.pl
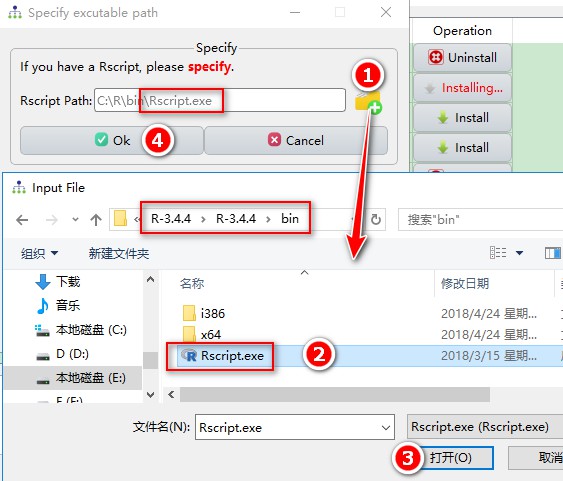
Rscript 3.4.4 | path-to-R/bin/Rscript.exe | path-to-R/bin/Rscript | path-to-R/bin/Rscript
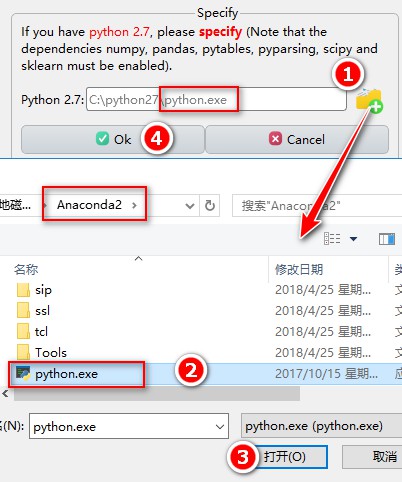
Python 2.7 | path-to-Python2.7/python.exe | path-to-Python2.7/bin/python | path-to-Python2.7/bin/python
tbl2asn | path-to-tbl2asn/win.tbl2asn/tbl2asn.exe | not allowed | not allowed
MPICH2 | not allowed | not allowed | path-to-MPICH2/bin/mpiexec or
path-to-MPICH2/bin/mpirun
2.2. Download plugins automatically
PhyloSuite also allows user to download the plugins and configure them automatically. Just click the Download button, and the plugin will be downloaded and installed on your computer automatically.

However, for Rscript and Python 2.7, there are two additional steps:
- When they are downloaded, you need to select a folder to install them;

- Next you need to specify the executable files, as described in Section 1.
Rscript:
Python 2.7:
In addition, if the plugin download fails, you can download and install them manually, via a link provided directly below (click “here”). Once downloaded, unzip them to your computer and specify the path.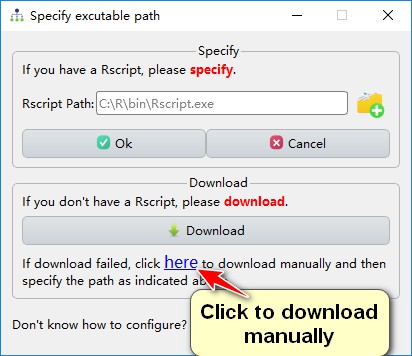
2.3. PartitionFinder configuration
As PartitionFinder2 Python script relies on Python 2.7, and additional six dependencies should be configured, we have compiled PartitionFinder2 (MAC and Windows only) so it does not rely on Python 2.7 any more.
- Open
PartitionFinder2configuration window; - Click
Downloadbutton in theInstall compiled PartitionFinder2box to download and install PartitionFinder2 automatically.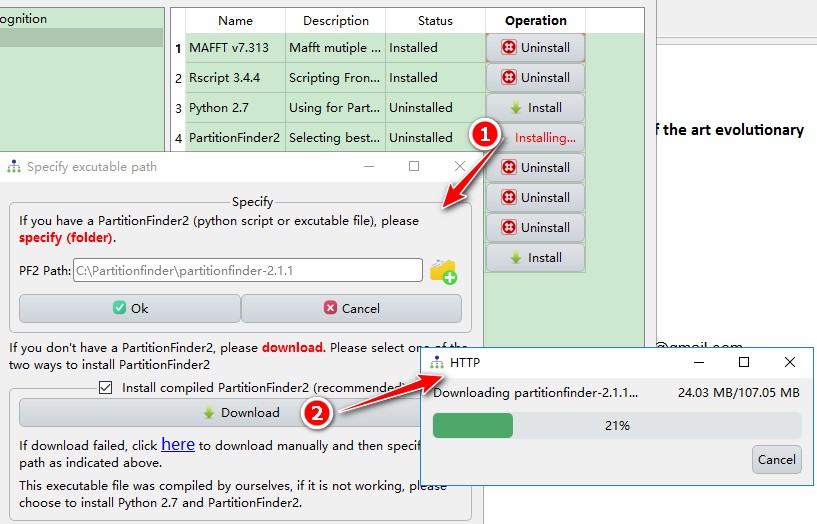
You may also select to install Python 2.7 and PartitionFinder2 Python script:
PartitionFinder2
- Open
PartitionFinder2configuration window; - Click
Downloadbutton in theInstall python 2.7 and PartitionFinder2box to download and install it automatically.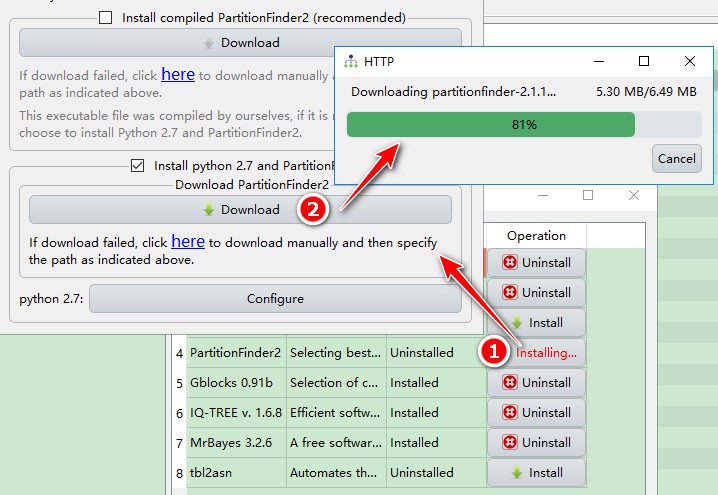
Python 2.7
- Open
Python 2.7configuration window; - Click
Downloadbutton in theDownloadbox to downloadAnaconda2. For the additional steps to complete the Python 2.7 configuration please refer to Section 2.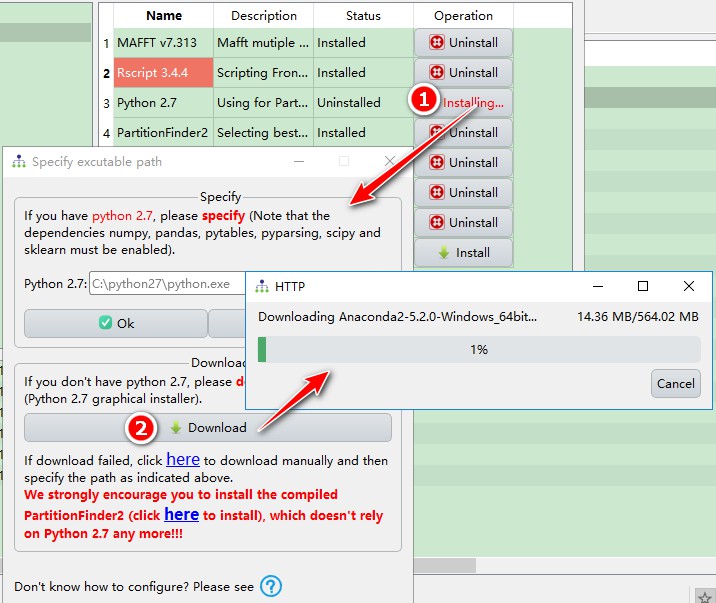
Note: Anaconda Python distribution will download for Python 2.7 (because it contains all of the dependencies required by PartitionFinder2: numpy, pandas, pytables, pyparsing, scipy and sklearn). As it is around 500M in size, your download may take some time. That’s why we recommend that you download the compiled PartitionFinder2, which does not require Python 2.7.
2.3.1. Troubleshooting
If you specify the path to Python 2.7 pre-installed on your computer, sometimes you may encounter the error below, indicating the absence of the six dependencies.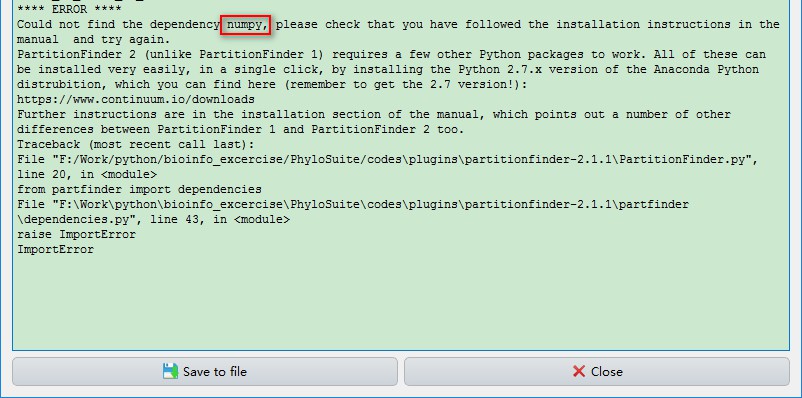
There are three solutions:
- Install compiled PartitionFinder2 (recommended, MAC and Windows only);
- Install
Anaconda2and specify thepythonexecutable file, as shown above; - Install these six dependencies to Python 2.7:
1
2
3
4
5
6pip install pandas
pip install numpy
pip install tables
pip install pyparsing
pip install scipy
pip install sklearn
2.4. HmmCleaner configuration
HmmCleaner relies on HMMER, Perl 5 and a lot of dependencies. Therefore, some users may have difficulty in configuring it. Here we make a brief tutorial for the configuration of HmmCleaner.
2.4.1. Configuration of HMMER
- If you already have conda installed, you can install HMMER via
conda install -c bioconda hmmerorconda install -c bioconda/label/cf201901 hmmercommand. - If you don’t have Conda, you need to download and install HMMER here: http://hmmer.org.
Please Note that all executables from HMMER have to be added in the environment variable ($PATH).
2.4.2. Configuration of Perl and dependencies installation
Here I recommend Perl version 5.18.0, as I experienced failure in installing dependencies in version 5.26.
When perl is installed, you can use cpanm to install HmmCleaner:
1 | cpan App::cpanminus |
Alternatively, you can also install HmmCleaner following this instruction: https://metacpan.org/source/ARNODF/Bio-MUST-Apps-HmmCleaner-0.180750/INSTALL
It may take some times to install, if it installs successfully, HmmCleaner.pl will be automatically add to the environment variables. Then you need to close and reopen PhyloSuite to see if it installed successfully (if you see Uninstall button, it means success).
